The Mystery of Life's Origin:
Reassessing Current Theories
CHAPTER 9
Specifying How Work Is To Be Done
In Chapter 7 we saw that the work necessary to polymerize DNA and protein
molecules from simple biomonomers could potentially be accomplished
by energy flow through the system. Still, we know that such energy flow
is a necessary but not sufficient condition for polymerization of the macromolecules
of life. Arranging a pile of bricks into the configuration of a house requires
work. One would hardly expect to accomplish this work with dynamite, however.
Not only must energy flow through the system, it must be coupled in some
specific way to the work to be done. This being so, we devoted Chapter 8
to identifying various components of work in typical polymerization reactions.
In reviewing those individual work components, one thing became clear. The
coupling of energy flow to the specific work requirements in the formation
of DNA and protein is particularly important since the required configurational
entropy work of coding is substantial.
Theoretical Models for the Origin of DNA and Protein
A mere appeal to open system thermodynamics does little good. What must
be done is to advance a workable theoretical model of how the available
energy can be coupled to do the required work. In this chapter various theoretical
models for the origin of DNA and protein will be evaluated. Specifically,
we will discuss how each proposes to couple the available energy to the
required work, particularly the configurational entropy work of coding.
Chance
Before the specified complexity of living systems began to be appreciated,
it was thought that, given enough time, "chance" would explain
the origin of living systems. In fact, most textbooks state that chance
is the basic explanation for the origin of life. For example, Lehninger
in his classic textbook Biochemistry states,
We now come to the critical moment in evolution in which the
first semblance of "life" appeared, through the chance association
of a number of abiotically formed macromolecular components, to yield a
unique system of greatly enhanced survival value.1
More recently the viability of "chance" as a mechanism for the
origin of life has been severely challenged.2
We are now ready to analyze the "chance" origin of life using
the approach developed in the last chapter. This view usually assumes that
energy flow through the system is capable of doing the chemical and the
thermal entropy work, while the configurational entropy work of both selecting
and coding is the fortuitous product of chance.
To illustrate, assume that we are trying to synthesize a protein containing
101 amino acids. In eq. 8-14 we estimated that the total free energy increase
(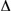 G) or work required to make a random polypeptide from previously
selected amino acids was 300 kcal/mole. An additional 159 kcal/mole is needed
to code the polypeptide into a protein. Since the "chance" model
assumes no coupling between energy flow and sequencing, the fraction of
the polypeptide that has the correct sequence may be calculated (eq. 8-16)
using equilibrium thermodynamics, i.e.,
G) or work required to make a random polypeptide from previously
selected amino acids was 300 kcal/mole. An additional 159 kcal/mole is needed
to code the polypeptide into a protein. Since the "chance" model
assumes no coupling between energy flow and sequencing, the fraction of
the polypeptide that has the correct sequence may be calculated (eq. 8-16)
using equilibrium thermodynamics, i.e.,
[protein concentration] / [polypeptide concentration] = exp (
- 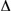 G / RT),
G / RT),  eq. (9-1)
eq. (9-1)
= exp (-159,000) / 1.9872 x 298)
or approximately 1 x 10-117
This ratio gives the fraction of polypeptides that have the right sequence
to be a protein.
[NOTE: This is essentially the inverse of the estimate for the
number of ways one can arrange 101 amino acids in a sequence (i.e., I /
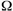 c
in eq. 8-7)].
c
in eq. 8-7)].
Eigen3 has estimated the number of polypeptides of molecular
weight 10 4 (the same weight used in our earlier calculations)
that would be found in a layer 1 meter thick covering the surface of the
entire earth. He found it to be 1041. If these polypeptides reformed
with new sequences at the maximum rate at which chemical reactions may occur,
namely 1014/s, for 5 x 109 years [1.6 x 1017
s], the total number of polypeptides that would be formed during the assumed
history of the earth would be
1041 x 1014/s x 1.6 x 1017s =
1072  (9-2)
(9-2)
Combining the results of eq. 9-1 and 9-2, we find the probability of
producing one protein of 101 amino acids in five billion years is only 1/
1045. Using somewhat different illustrations, Steinman4
and Cairns-Smith5 also come to the conclusion that chance
is insufficient.
It is apparent that "chance" should be abandoned as an acceptable
model for coding of the macromolecules essential in living systems. In fact,
it has been, except in introductory texts and popularizations.
Neo-Darwinian Natural Selection
The widespread recognition of the severe improbability that self-replicating
organisms could have formed from purely random interactions has led to a
great deal of speculation---speculation that some organizing principle must
have been involved. In the company of many others, Crick6 has
considered that the neo-Darwinian mechanism of natural selection might provide
the answer. An entity capable of self-replication is necessary, however,
before natural selection can operate. Only then could changes result via
mutations and environmental pressures which might in turn bring about the
dominance of entities with the greatest probabilities of survival and reproduction.
The weakest point in this explanation of life's origin is the great complexity
of the initial entity which must form, apparently by random fluctuations,
before natural selection can take over. In essence this theory postulates
the chance formation of the "metabolic motor" which will subsequently
be capable of channeling energy flow through the system. Thus harnessed
by coupling through the metabolic motor, the energy flow is imagined to
supply not only chemical and thermal entropy work, but also the configurational
entropy work of selecting the appropriate chemicals and then coding the
resultant polymer into an aperiodic, specified, biofunctioning polymer.
As a minimum, this system must carry in its structure the information for
its own synthesis, and control the machinery which will fabricate any desired
copy. It is widely agreed that such a system requires both protein and nucleic
acid.7 This view is not unanimous, however. A few have suggested
that a short peptide would be sufficient.8
One way out of the problem would be to extend the concept of natural selection
to the pre-living world of molecules. A number of authors have entertained
this possibility, although no reasonable explanation has made the suggestion
plausible. Natural selection is a recognized principle of differential reproduction
which presupposes the existence of at least two distinct types of self-replicating
molecules. Dobzhansky appealed to those doing origin-of-life research not
to tamper with the definition of natural selection when he said:
I would like to plead with you, simply, please realize you cannot
use the words "natural selection" loosely. Prebiological natural
selection is a contradiction in terms.9
Bertalanffy made the point even more cogently:
Selection, i.e., favored survival of "better" precursors
of life, already presupposes self-maintaining, complex, open systems which
may compete; therefore selection cannot account for the origin of such systems.10
Inherent Self-Ordering Tendencies in Matter
How could energy flow through the system be sufficiently coupled to do the
chemical and thermal entropy work to form a nontrivial yield of polypeptides
(as previously assumed in the "chance" model)? One answer has
been the suggestion that configurational entropy work, especially the coding
work, could occur as a consequence of the self-ordering tendencies in matter.
The experimental work of Steinman and Cole11 in the late Sixties
is still widely cited in support of this model.12 The polymerization
of protein is hypothesized to be a nonrandom process, the coding
of the protein resulting from differences in the chemical bonding forces.
For example, if amino acids A and B react chemically with one another more
readily than with amino acids C, D, and E, we should expect to see a greater
frequency of AB peptide bonds in protein than AC, AD, AE, or BC, BD, BE
bonds.
Together with our colleague Randall Kok, we have recently analyzed the ten
proteins originally analyzed by Steinman and Cole,13 as well
as fifteen additional proteins whose structures (except for hemoglobin)
have been determined since their work was first published in 1967. Our expectation
in this study was that one would only get agreement between the dipeptide
bond frequencies from Steinman and Cole's work and those observed in actual
proteins if one considered a large number of proteins averaged together.
The distinctive structures of individual proteins would cause them to vary
greatly from Steinman and Cole's data, so only when these distinctives are
averaged out could one expect to approach Steinman and Cole's dipeptide
bond frequency results. The reduced data presented in table 9-1 shows that
Steinman and Cole's dipeptide bond frequencies do not correlate well with
the observed peptide bond frequencies for one, ten, or twenty-five proteins.
It is a simple matter to make such calculations on an electronic digital
computer. We surmise that additional assumptions not stated in their paper
were used to achieve the better agreements.
Furthermore, the peptide bond frequencies for the twenty-five proteins approach
a distribution predicted by random statistics rather than the dipeptide
bond frequency measured by Steinman and Cole. This observation means that
bonding preferences between various amino acids play no significant role
in coding protein. Finally, if chemical bonding forces were influential
in amino acid sequencing, one would expect to get a single sequence (as
in ice crystals) or no more than a few sequences, instead of the large variety
we observe in living systems. Yockey, with a different analysis, comes to
essentially the same conclusion.14
A similar conclusion may be drawn for DNA synthesis. No one to date has
published data indicating that bonding preferences could have had any role
in coding the DNA molecules. Chemical bonding forces apparently have minimal
effect on the sequence of nucleotides in a polynucleotide.
Table 9-1.
Comparison of Steinman and Cole's experimentally determined dipeptide bond
frequencies, and frequencies calculated by Steinman and Cole, and by Kok
and Bradley from known protein sequences.
Dipeptide*
|
Values (relative to Gly-Gly)
| | | |
| | |
| |
|
S / C+
|
K / B #
| | |
|
exp &
|
cal
|
cal-wa
|
cal-woa
|
Gly-Gly
|
1.0
|
1.0
|
1.0 (1.0) [1.0]
|
1.0 (1.0) [1.0]
|
Gly-Ala
|
0.8
|
0.7
|
1.1 (1.1) [2.0]
|
2.0 (1.2) [1.0]
|
Ala-Gly
|
0.8
|
0.6
|
1.0 (1.1) [2.2]
|
1.5 (1.2) [0.0]
|
Ala-Ala
|
0.7
|
0.6
|
1.3 (1.5) [4.4]
|
2.8 (1.5) [0.0]
|
Gly-Val
|
0.5
|
0.2
|
0.2 (0.3) [0.4]
|
1.5 (1.2) [1.0]
|
Val-Gly
|
0.5
|
0.3
|
0.3 (0.3) [0.6]
|
0.8 (0.6) [0.0]
|
Gly-Leu
|
0.5
|
0.3
|
0.3 (0.3) [0.2]
|
1.3 (0.7) [1.0]
|
Leu-Gly
|
0.5
|
0.2
|
0.3 (0.3) [0.8]
|
1.3 (1.0) [1.0]
|
Gly-Ile
|
0.3
|
0.1
|
0.1 (0.2) [0.6]
|
1.0 (0.8) [0.0]
|
Ile-Gly
|
0.3
|
0.1
|
0.1 (0.2) [0.2]
|
0.0 (0.4) [0.0]
|
Gly-Phe
|
0.1
|
0.1
|
0.1 (0.2) [0.4]
|
0.5 (0.5) [0.0]
|
Phe-Gly
|
0.1
|
0.1
|
0.1 (0.1) [0.6]
|
1.0 (0.5) [1.0]
|
(Adapted after G. Steinman and M.V. Cole, 1967. Proc. Nat. Acad.
Sci. U.S. 58,735).
* The dipeptides are listed in terms of increasing volume of the side chains
of the constituent residues. Gly = glycine, Ala = alanine, Val = valine,
Leu = leucine, Ile = isoleucine and Phe = phenylalanine. Example: Gly-Ala
= glycylalanine.
+ Steinman and Cole's (S/C) experimentally determined dipeptide bond frequencies
were normalized and compared to the calculated frequencies obtained by counting
actual peptide bond frequencies in ten proteins, assuming all seryl and
threonyl residues are counted as glycine and all aspartyl and glutamyl residues
are counted as alanine. The ten proteins used were: egg lysozyme, ribonuclease,
sheep insulin, whale myoglobin, yeast cytochrome c, tobacco mosaic virus,
beta-corticotropin, glucagon, melanocyte-stimulating hormone, and chymotrypsinogen.
Because of ambiguity regarding sequences used by S/C, all sequences are
those shown in Atlas of Protein Sequence and Structure, 1972.
Vol. V (ed. by M.O. Dayhoff). National Biomedical Research Foundation, Georgetown
University Medical Center, Washington, D.C.
& The experimentally determined dipeptide frequencies were obtained with
aqueous solutions containing 0.01 M each amino acid, 0.125 N HCl, 0.1 M
sodium dicyanamide.
#Kok and Bradley's (K/Bcalculated dipeptide frequencies were obtained by
counting S?Cassumptions. The numbers in brackets are for one protein, enterotoxin
B, with actual peptide bond frequencies for the same ten proteins with (wa)
and without (woa) S/C assumptions. The numbers in parentheses are for twenty-five
proteins with (wa) and without (woa) S/C assumptions. The twenty-five proteins
are the ten used S/C and alpha S1 Casein (bovine); azurin (bordetella bronchisetica);
carboxypeptidase A (bovine); cytochrome b5 (bovine); enterotoxin B; elastase
(pig); glyceraldehyde 3-phosphate dehydrogenase (lobster); human growth
hormone; human hemoglobin beta chain; histone 11B2 (bovine); immunoglobulin
gamma-chain 1, V-I (human EU); penicillinase (bacillus licheniformis 749/c);
sheep prolactin; subtilisin (bacillus amyloliquefaciens); and tryptophan
synthetase alpha chain (E-coh K-i 2). Sequences are those shown
in Atlas of Protein Sequence and Structure, 1972. Vol. V (ed.
by M.O. Dayhoff). Note disagreement S/C K/B calculated results. Also S/C
calculated results are at variance with S/C experimental values for one,
ten or twenty-five proteins, with (wa) or without (woa) S/C assumptions.
Mineral Catalysis
Mineral catalysis is often suggested as being significant in prebiotic evolution.
In the experimental investigations reported in the early 1970's15
mineral catalysis in polymerization reactions was found to operate by adsorption
of biomonomers on the surface or between layers of clay. Monomers were effectively
concentrated and protected from rehydration so that condensation polymerization
could occur. There does not appear to be any additional effect. In considering
this catalytic effect of clay, Hulett has advised, "It must be remembered
that the surface cannot change the free energy relationships between reactants
and products, but only the speed with which equilibrium is reached."16
Is mineral catalysis capable of doing the chemical work and/or thermal entropy
work? The answer is a qualified no. While it should assist in doing the
thermal entropy work, it is incapable of doing the chemical work since clays
do not supply energy. This is why successful mineral catalysis experiments
invariably use energy-rich precursors such as aminoacyl adenylates rather
than amino acids.17
Is there a real prospect that mineral catalysis may somehow accomplish the
configurational entropy work, particularly the coding of polypeptides or
polynucleotides? Here the answer is clearly no. In all experimental work
to date, only random polymers have been condensed from solutions of selected
ingredients. Furthermore, there is no theoretical basis for the notion that
mineral catalysis could impart any significant degree of information content
to polypeptides or polynucleotides. As has been noted by Wilder-Smith,18
there is really no reason to expect the low-grade order resident on minerals
to impart any high degree of coding to polymers that condense while adsorbed
on the mineral's surface. To put it another way, one cannot get a complex,
aperiodic-sequenced polymer using a very periodic (or crystalline) template.
In summary, mineral catalysis must be rejected as a mechanism for doing
either the chemical or configurational entropy work required to polymerize
the macromolecules of life. It can only assist in polymerizing short, random
chains of polymers from selected high-energy biomonomers by assisting in
doing the thermal entropy work.
Nonlinear, Nonequilibrium Processes
1. Ilya Prigogine
Prigogine has developed a more general formulation of the laws of thermodynamics
which includes nonlinear, irreversible processes such as autocatalytic activity.
In his book Self Organization in Nonequilibrium Systems (1977)19
co-authored with Nicolis, he summarized this work and its application to
the organization and maintenance of highly complex structures in living
things. The basic thesis in the book is that there are some systems which
obey non-linear laws---laws that produce two distinct kinds of behavior.
In the neighborhood of thermodynamic equilibrium, destruction of order prevails
(entropy achieves a maximum value consistent with the system constraints).
If these same systems are driven sufficiently far from equilibrium, however,
ordering may appear spontaneously.
Heat flow by convection is an example of this type of behavior. Heat conduction
in gases normally occurs by the random collision of gas molecules. Under
certain conditions, however, heat conduction may occur by a heat-convection
current---the coordinated movement of many gas molecules. In a similar way,
water flow out of a bathtub may occur by random movement of the water molecules
under the influence of gravity. Under certain conditions, however, this
random movement of water down the drain is replaced by the familiar soapy
swirl---the highly coordinated flow of the vortex. In each case random movements
of molecules in a fluid are spontaneously replaced by a highly ordered behavior.
Prigogine et al.,20 Eigen,21 and others have
suggested that a similar sort of self-organization may be intrinsic in organic
chemistry and can potentially account for the highly complex macromolecules
essential for living systems.
But such analogies have scant relevance to the origin-of-life question.
A major reason is that they fail to distinguish between order and complexity.
The highly ordered movement of energy through a system as in convection
or vortices suffers from the same shortcoming as the analogies to the static,
periodic order of crystals. Regularity or order cannot serve to store the
large amount of information required by living systems. A highly irregular,
but specified, structure is required rather than an ordered structure. This
is a serious flaw in the analogy offered. There is no apparent connection
between the kind of spontaneous ordering that occurs from energy flow through
such systems and the work required to build aperiodic information-intensive
macromolecules like DNA and protein. Prigogine, et al.22
suggest that the energy flow through the system decreases the system entropy,
leading potentially to the highly organized structure of DNA and protein.
Yet they offer no suggestion as to how the decrease in thermal entropy from
energy flow through the system could be coupled to do the configurational
entropy work required.
A second reason for skepticism about the relevance of the models developed
by Prigogine, et al.23 and others is that ordering produced
within the system arises through constraints imposed in an implicit way
at the system boundary. Thus, the system order, and more importantly the
system complexity, cannot exceed that of the environment.
Walton24 illustrates this concept in the following way. A container
of gas placed in contact with a heat source on one side and a heat sink
on the opposite side is an open system. The flow of energy through the system
from the heat source to the heat sink forms a concentration relative to
the gas in the cooler region. The order in this system is established by
the structure: source-intermediate systems-sink. If this structure is removed,
allowing the heat source to come into contact with the heat sink, the system
decays back to equilibrium. We should note that the information induced
in an open system doesn't exceed the amount of information built into the
structural environment, which is its source.
Condensation of nucleotides to give polynucleotides or nucleic acids can
be brought about with the appropriate apparatus (i.e., structure) and supplies
of energy and matter. Just as in Walton's illustration, however, Mora25has
shown that the amount of order (not to mention specified complexity) in
the final product is no greater than the amount of information introduced
in the physical structure of the experiment or chemical structure of the
reactants. Non-equilibrium thermodynamics does not account for this structure,
but assumes it and then shows the kind of organization which it produces.
The origin and maintenance of the structure are not explained, and as Harrison26
correctly notes this question leads back to the origin of structure in the
universe. Science offers us no satisfactory answer to this problem at present.
Nicolis and Prigogine27 offer their trimolecular model as an
example of a chemical system with the required nonlinearity to produce self
ordering. They are able to demonstrate mathematically that within a system
that was initially homogeneous, one may subsequently have a periodic, spatial
variation of concentration. To achieve this low degree of ordering, however,
they must require boundary conditions that could only be met at cell walls
(i.e., at membranes), relative reaction rates that are atypical of those
observed in condensation reactions, a rapid removal of reaction flow products,
and a trimolecular reaction (the highly unlikely simultaneous collision
of three atoms). Furthermore the trimolecular model requires chemical reactions
that are essentially irreversible. But condensation reactions for polypeptides
or polynucleotides are highly reversible unless all water is removed from
the system.
They speculate that the low degree of spatial ordering achieved in the simple
trimolecular model could potentially be orders of magnitude greater for
the more complex reactions one might observe leading up to a fully replicating
cell. The list of boundary constraints, relative reaction rates, etc. would,
however, also be orders of magnitude larger. As a matter of fact, one is
left with so constraining the system at the boundaries that ordering is
inevitable from the structuring of the environment by the chemist. The fortuitous
satisfaction of all of these boundary constraints simultaneously would be
a its miracle in its own right.
It is possible at present to synthesize a few proteins such as insulin in
the laboratory. The chemist supplies not only energy to do the chemical
and thermal entropy work, however, but also the necessary chemical manipulations
to accomplish the configurational entropy work. Without this, the selection
of the proper composition and the coding for the right sequence of amino
acids would not occur. The success of the experiment is fundamentally dependent
on the chemist.
Finally, Nicolis and Prigogine have postulated that a system of chemical
reactions which explicitly shows autocatalytic activity may ultimately be
able to circumvent the problems now associated with synthesis of prebiotic
DNA and protein. It remains to be demonstrated experimentally, however,
that these models have any real correspondence to prebiotic condensation
reactions. At best, these models predict higher yields without any mechanism
to control sequencing. Accordingly, no experimental evidence has been reported
to show how such models could have produced any significant degree of coding.
No, the models of Prigogine et al., based on non-equilibrium thermodynamics,
do not at present offer an explanation as to how the configurational entropy
work is accomplished under prebiotic conditions. The problem of how to couple
energy flow through the system to do the required configurational entropy
work remains.
2. Manfred Eigen
In his comprehensive application of nonequilibrium thermodynamics to the
evolution of biological systems, Eigen28 has shown that selection
could produce no evolutionary development in an open system unless the system
were maintained far from equilibrium. The reaction must be autocatalytic
but capable of self-replication. He develops an argument to show that in
order to produce a truly self-replicating system the complementary base-pairing
instruction potential of nucleic acids must be combined with the catalytic
coupling function of proteins. Kaplan29 has suggested a minimum
of 20-40 functional proteins of 70-100 amino acids each, and a similar number
of nucleic acids would be required by such a system. Yet as has previously
been noted, the chance origin of even one protein of 100 amino acids is
essentially zero.
The shortcoming of this model is the same as for those previously discussed;
namely, no way is presented to couple the energy flow through the system
to achieve the configurational entropy work required to create a system
capable of replicating itself.
Periodically we see reversions (perhaps inadvertent ones) to chance in the
theoretical models advanced to solve the problem. Eigen's model illustrates
this well. The model he sets forth must necessarily arise from chance events
and is nearly as incredible as the chance origin of life itself. The fact
that generally chance has to be invoked many times in the abiotic sequence
has been called by Brooks and Shaw "a major weakness in the whole chemical
evolutionary theory."30
Experimental Results in Synthesis of Protein and DNA
Thus far we have reviewed the various theoretical models proposed to explain
how energy flow through a system might accomplish the work of synthesizing
protein and DNA macromolecules, but found them wanting. Nevertheless, it
is conceivable that experimental Support for a spontaneous origin of life
can be found in advance of the theoretical explanation for how this occurs.
What then can be said of the experimental efforts to synthesize protein
and DNA macromolecules? Experimental efforts to this end have been enthusiastically
pursued for the past thirty years. In this section, we will review efforts
toward the prebiotic syntheses of both protein and DNA, considering the
three forms of energy flow most commonly thought to have been available
on the early earth. These are thermal energy (volcanoes), radiant energy
(sun), and chemical energy in the form of either condensing agents or energy-rich
precursors. (Electrical energy is excluded at this stage of evolution as
being too "violent," destroying rather than joining the biomonomers.)
Thermal Synthesis
Sidney Fox31 has pioneered the thermal synthesis of polypeptides,
naming the products of his synthesis proteinoids. Beginning with
either an aqueous solution of amino acids or dry ones, he heats his material
at 2000oC for 6-7 hours.
[NOTE: Fox has modified this picture in recent years by developing
"low temperature" syntheses, i.e., 90-120oC. See S.
Fox, 1976. J Mol Evol 8, 301; and D. Rohlfing, 1976.
Science 193, 68].
All initial solvent water, plus water produced during Polymerization, is
effectively eliminated through vaporization. This elimination of the water
makes possible a small but significant yield of polypeptides, some with
as many as 200 amino acid units. Heat is introduced into the system by conduction
and convection and leaves in the form of steam. The reason for the success
of the polypeptide formation is readily seen by examining again equations
8-15 and 8-16. Note that increasing the temperature would increase the product
yield through increasing the value of exp (- 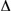 G / RT. But more importantly,
eliminating the water makes the reaction irreversible, giving an enormous
increase in yield over that observed under equilibrium conditions by the
application of the law of mass action.
G / RT. But more importantly,
eliminating the water makes the reaction irreversible, giving an enormous
increase in yield over that observed under equilibrium conditions by the
application of the law of mass action.
Thermal syntheses of polypeptides fail, however, for at least four reasons.
First, studies using nuclear magnetic resonance (NMR) have shown that thermal
proteinoids "have scarce resemblance to natural peptidic material because
beta, gamma, and epsilon peptide bonds largely predominate over alpha-peptide
bonds."32
[NOTE: This quotation refers to peptide links involving the
beta-carboxyl group of aspartic acid, the gamma-carboxyl group of glutamic
acid, and the epsilon-amino group of lysine which are never found in natural
proteins. Natural proteins use alpha-peptide bonds exclusively].
Second, thermal proteinoids are composed of approximately equal numbers
of L- and D-amino acids in contrast to viable proteins with all L-amino
acids. Third, there is no evidence that proteinoids differ significantly
from a random sequence of amino acids, with little or no catalytic activity.
[It is noted, however, that Fox has long disputed this.] Miller and Orgel
have made the following observation with regard to Fox's claim that proteinoids
resemble proteins:
The degree of nonrandomness in thermal polypeptides so far demonstrated
is minute compared to nonrandomness of proteins. It is deceptive, then,
to suggest that thermal polypeptides are similar to proteins in their nonrandomness.33
Fourth, the geological conditions indicated are too unreasonable to be taken
seriously. As Folsome has commented, "The central question [concerning
Fox's proteinoids] is where did all those pure, dry, concentrated, and optically
active amino acids come from in the real, abiological world?"34
There is no question that thermal energy flow through the system including
the removal of water is accomplishing the thermal entropy and chemical work
required to form a polypeptide (300 kcal/mole in our earlier example). The
fact that polypeptides are formed is evidence of the work done. It is equally
clear that the additional configurational entropy work required to convert
an aperiodic unspecified polypeptide into a specified, aperiodic polypeptide
which is a functional protein has not been done (159 kcal/mole in our earlier
example).
It should be remembered that this 159 kcal/mole of configurational entropy
work was calculated assuming the sequencing of the amino acids was the only
additional work to be done. Yet the experimental results of Temussi et
al.,35 indicate that obtaining all Lamino acids from a racemic
mixture and getting alpha-linking between the amino acids are quite difficult.
This requirement further increases the configurational entropy work needed
over that estimated to do the coding work (159 kcal/mole). We may estimate
the magnitude of this increase in the configurational entropy work term
by returning to our original calculations (eq. 8-7 and 8-8).
In our original calculation for a hypothetical protein of 100 amino acid
units, we assumed the amino acids were equally divided among the twenty
types. We calculated the number of possible amino acid sequences as follows:
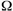 cr = 100! / 5! 5! 5!....5! = 100! / (5!)20
= 1.28 x 10115
cr = 100! / 5! 5! 5!....5! = 100! / (5!)20
= 1.28 x 10115  (9-3)
(9-3)
If we note that at each site the probability of having an L-amino acid
is 50%, and make the generous assumption that there is a 50% probability
that a given link will be of the alpha-type observed in true proteins, then
the number of ways the system can be arranged in a random chemical reaction
is given by
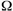 cr = 1.28 x 10115 x 2100
x 299 = 10175
cr = 1.28 x 10115 x 2100
x 299 = 10175  (9-4)
(9-4)
where 2100 refers to the number of additional arrangements
possible, given that each site could contain an L- or D-amino acid, and
299 assumes the 99 links between the 100 amino acids in general
are equally divided between the natural alpha-links and the unnatural beta-,
gamma-, or epsilon-links.
[NOTE Some studies indicate less than 50% alpha-links in peptides
formed by reacting random mixtures of amino acids. (P.A. Temussi, L. Paolillo,
F.E. Benedetti, and S. Andini, 1976. J. Mol. Evol. 7, 105.)].
The requirements for a biologically functional protein molecule are: (1)
all L-amino acids, (2) all alpha-links, and (3) a specified sequence. This
being so, the calculation of the configurational entropy of the protein
molecule using equation 8-8 is unchanged except that the number of ways
the system can be arranged, (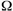 cr), is increased from 1.28
x 10115 to 1.0 x 10175 as shown in equations 9-3 and
9-4. We may use the relationships of equations 8-7 and 8-8 but with the
number of permutations modified as shown here to find a total configurational
entropy work. When we do, we get a total configurational entropy work of
195 kcal/mole, of which 159 kcal/mole is for sequencing and 36 kcal/mole
to attain all L-amino acids and all alpha-links. Finally, it should be recognized
that Fox and others who use his approach avoid a much larger configurational
entropy work term by beginning with only amino acids, i.e., excluding other
organic chemicals and thereby eliminating the "selecting work"
which is not accounted for in the 195 kcal/mole calculated above.
cr), is increased from 1.28
x 10115 to 1.0 x 10175 as shown in equations 9-3 and
9-4. We may use the relationships of equations 8-7 and 8-8 but with the
number of permutations modified as shown here to find a total configurational
entropy work. When we do, we get a total configurational entropy work of
195 kcal/mole, of which 159 kcal/mole is for sequencing and 36 kcal/mole
to attain all L-amino acids and all alpha-links. Finally, it should be recognized
that Fox and others who use his approach avoid a much larger configurational
entropy work term by beginning with only amino acids, i.e., excluding other
organic chemicals and thereby eliminating the "selecting work"
which is not accounted for in the 195 kcal/mole calculated above.
In summary, undirected thermal energy is only able to do the chemical and
thermal entropy work in polypeptide synthesis, but not the coding (or sequencing)
portion of the configurational entropy work. Protenoids are just globs of
random polymers. That a polymer composed exclusively of amino acids (but
without exclusively peptide bonds) was formed is a result of the fact that
only amino acids were used in the experiment. Thus, the portion of the configurational
entropy work that was done---the selecting work---was accomplished not by
natural forces but by illegitimate investigator interference. It is difficult
to imagine how one could ever couple random thermal energy flow through
the system to do the required configurational entropy work of selecting
and sequencing. Finally, this approach is of very questionable geological
significance, given the many fortuitous events that are required, as others
have noted.
Solar Energy
Direct photochemical (UV) polymerization reactions to form polypeptides
and polynucleotides have occasionally been discussed in the literature.
The idea is to drive forward the otherwise thermodynamically unfavorable
polymerization reaction by allowing solar energy to flow through the aqueous
system to do the necessary work. It is worth noting that minor yields of
small peptides can be expected to form spontaneously, even though the reaction
is unfavorable (see eq. 8-16), but that greater yields of larger peptides
can be expected only if energy is somehow coupled to the reaction. Fox and
Dose have examined the peptide results of Bahadur and Ranganayaki36
and concluded that UV irradiation did not couple with the reaction. They
comment, "The authors do not show that they have done more than accelerate
an approach to an unfavorable equilibrium. They may merely have reaffirmed
the second law of thermodynamics."37 Other attempts to form
polymers directly under the influence of UV light have not been encouraging
because of this lack of coupling. Neither the chemical nor the thermal entropy
work, and definitely not any configurational entropy work, has been accomplished
using solar energy.
Chemical Energy (Energy-Rich Condensing Agents)
Through the use of condensing agents, the energetically unfavorable dipeptide
reaction (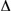 G1 = + 3000 cal/mole) is made energetically favorable
(
G1 = + 3000 cal/mole) is made energetically favorable
(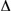 G3 < 0) by coupling it with a second reaction which is
sufficiently favorable energetically (
G3 < 0) by coupling it with a second reaction which is
sufficiently favorable energetically (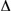 G2 < 0), to offset the
energy requirement of the dipeptide reaction:
G2 < 0), to offset the
energy requirement of the dipeptide reaction:
dipeptide reaction
A - OH + H - B 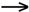 A - B + H20
A - B + H20
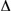 G1 > 0
G1 > 0
 (9-5)
(9-5)
condensing agent reaction
C + H20 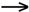 D
D
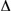 G2 < 0
G2 < 0  (9-6)
(9-6)
coupled reaction
A - OH + H - B + C 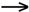 A - B + D
A - B + D
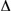 G3
< 0
G3
< 0  (9-7)
(9-7)
As in thermal proteinoid formation, the free water is removed. However,
in this case, it is removed by chemical reaction with a suitable poly- condensing
agent-one which has a sufficient decrease in Gibbs free energy to drive
the reaction forward (i.e., 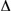 G2 0 and |
G2 0 and | 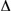 G2
|
G2
| 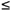 |G1 | so that
|G1 | so that 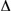 G1 +
G1 + 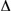 G2
=
G2
= 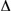 G3
G3 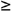 0.
0.
Unfortunately, it has proved difficult to find condensing agents work. for
these macromolecule syntheses that could have originated on the primitive
earth and functioned properly under mild conditions in an aqueous environment.38
Meanwhile, other condensing agents which are not prebiotically significant
(e.g., polymetaphosphates) are used in experiments. The plausible cyanide
derivative candidates for condensing agents on the early earth hydrolyze
readily in aqueous solutions (see Chapter 4). In the process, they do not
couple preferentially with the H20 from the condensation-dehydration
reaction. Condensing agents observed in living systems today are produced
only by living systems, and thus are not prebiotically significant. Moreover,
enzyme activity in living systems first activates amino acids and then brings
about condensation of these activated species, thus avoiding the problem
of indiscriminate reaction with water.
Notice that if we could solve the very significant problems associated with
the prebiotic synthesis of polypeptides by using condensing agents, we would
still succeed only in polymerizing random polypeptides. Only the chemical
and thermal entropy work would be accomplished by an appropriate coupling
of the condensing agent to the condensation reaction. There is no reason
to believe that condensing agents could have any effect on the selecting
or sequencing of the amino acids. Thus, condensing agents are eliminated
as a possible means of doing the configurational entropy work of coding
a protein or DNA.
Chemical Energy (Energy-Rich Precursors)
Because the formation of even random polypeptides from amino acids is so
energetically unfavorable (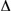 G = 300 kcal/mole for 100 amino acids),
some investigators have attempted to begin with energy-rich precursors such
as HCN and form polypeptides directly, a scheme which is "downhill"
energetically, i.e.,
G = 300 kcal/mole for 100 amino acids),
some investigators have attempted to begin with energy-rich precursors such
as HCN and form polypeptides directly, a scheme which is "downhill"
energetically, i.e., 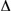 G < 0. There are advantages to such an approach; namely,
there is no chemical work to be done since the bonding energy actually decreases
as the energy-rich precursors react to form more complex molecules. This
decrease in bonding energy will drive the reaction forward, effectively
doing the thermal entropy work as well. The fly in the ointment, however,
is that the configurational entropy work is enormous in going from simple
molecules (e.g., HCN) directly to complex polymers in a single step (without
forming intermediate biomonomers).
G < 0. There are advantages to such an approach; namely,
there is no chemical work to be done since the bonding energy actually decreases
as the energy-rich precursors react to form more complex molecules. This
decrease in bonding energy will drive the reaction forward, effectively
doing the thermal entropy work as well. The fly in the ointment, however,
is that the configurational entropy work is enormous in going from simple
molecules (e.g., HCN) directly to complex polymers in a single step (without
forming intermediate biomonomers).
The stepwise scheme of experiments is to react gases such as methane, ammonia,
and carbon dioxide to form amino acids and other compounds and then to react
these to form polymers in a subsequent experiment. In these experiments
the very considerable selecting-work component of the configurational entropy
work is essentially done by the investigator who separates, purifies, and
concentrates the amino acids before attempting to polymerize them. Matthews39
and co-workers, however, have undertaken experiments where this intermediate
step is missing and the investigator has no opportunity to contribute even
obliquely to the success of the experiment by assisting in doing the selecting
part of the configurational entropy work. In such experiments-undoubtedly
more plausible as true prebiotic simulations-the probability of success
is, however, further reduced from the already small probabilities previously
mentioned. Using HCN as an energy-rich precursor, and ammonia as a catalyst,
Matthews and Moser40 have claimed direct synthesis of a large
variety of chemicals under anhydrous conditions. After treating the polymer
with water, even peptides are said to be among the products obtained. But
as Ferris et al.,41 have shown, the HCN polymer does not
release amino acids upon treatment with proteolytic (protein splitting)
enzymes; nor does it give a positive biuret reaction (color test for peptides).
In short, it is very hard to reconcile these results with a peptidic structure.
Ferris42 and Matthews43 have agreed that direct synthesis
of polypeptides has not yet been demonstrated. While some peptide bonds
may form directly, it would be quite surprising to find them in significant
numbers. Since HCN gives rise to other organic compounds, and various kinds
of links are possible, the formation of polypeptides with exclusively alpha-links
is most unlikely. Furthermore, no sequencing would be expected from this
reaction, which is driven forward and "guided" only by chemical
energy.
While we do not believe Matthews or others will be successful in demonstrating
a single step synthesis of polypeptides from HCN, this approach does involve
the least investigator interference, and thus, represents a very plausible
prebiotic simulation experiment. The approach of Fox and others, which involves
reacting gases to form many organic compounds, separating out amino acids,
purifying, and finally polymerizing them, is more successful because it
involves a greater measure of investigator interference. The selecting portion
of the configurational entropy work is being supplied by the scientist.
Matthew's lack of demonstrable success in producing polypeptides is a predictable
indication of the enormity of the problem of prebiotic synthesis when it
is not overcome by illegitimate investigator interference.
Mineral Catalysis
A novel synthesis of polypeptides has been reported44 which employs
mineral catalysis. An aqueous solution of energy-rich aminoacyl adenylates
(rather than amino acids) is used in the presence of certain layered clays
such as those known as montmorillonites. Large amounts of the energy-rich
reactants are adsorbed both on the surface and between the layers of clay.
The catalytic effect of the clay may result primarily from the removal of
reactants from the solution by adsorption between the layers of clay. This
technique has resulted in polypeptides of up to 50 units or more. Although
polymerization definitely occurs in these reactions, the energy-rich aminoacyl
adenylate (fig. 9-1) is of very doubtful prebiotic significance per the
discussion of competing reactions in Chapter 4. Furthermore, the use of
clay with free amino acids will not give a successful synthesis of polypeptides.
The energy-rich aminoacyl adenylates lower their chemical or bonding energy
as they polymerize, driving the reaction forward, and effectively doing
the thermal entropy work as well. The role of the clay is to concentrate
the reactants and possibly to catalyze the reactions. Once again, we are
left with no apparent means to couple the energy flow, in this case in the
form of prebiotically questionable energy-rich precursors, to the configurational
entropy work of selecting and sequencing required in the formation of specified
aperiodic polypeptides, or proteins.
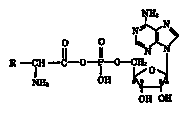
Figure 9-1.
Aminoacyl adenylate.
Summary of Experimental Results on Prebiotic Synthesis of protein
In summary, we have seen that it is possible to do the thermal entropy work
and chemical work necessary to form random polypeptides, e.g., Fox's proteinoids.
In no case, though, has anyone been successful in doing the additional configurational
entropy work of coding necessary to convert random polypeptides into proteins.
Virtually no mechanism with any promise for coupling the random flow of
energy through the system to do this very specific work has come to light.
The prebiotic plausibility of the successful synthesis of polypeptides must
be questioned because of the considerable configurational entropy work of
selecting done by the investigator prior to the polymer synthesis. Surely
no suggestion is forthcoming that the right composition of just the subset
of amino acids found in living things was "selected" by natural
means, or that this subset consists only of L-a-amino acids. This is precisely
why a large measure of the credit in forming proteinoids must go to Fox
and others rather than nature.
Summary of Experimental Results on Prebiotic Synthesis of DNA
The prebiotic synthesis of DNA has proved to be even more difficult than
that of protein. The problems that beset protein synthesis apply with greater
force to DNA synthesis. Energy flow through the system may cause the nucleotides
to chemically react and form a polymer chain, but it is very difficult to
get them to attach themselves together in a specified way. For example,
3' - 5' links on the sugar are necessary for the DNA to form a helical structure
(see fig. 9-2). Yet 2'-5' links predominate in most prebiotic simulation
experiments.45 The sequencing of the bases in DNA is also crucial,
as is the amino acid sequence in proteins. Both of these requirements are
problems in doing the configurational entropy work. It is one thing to get
molecules to chemically react; it is quite another to get them to link up
in the right arrangement. To date, researchers have only succeeded in making
oligonucleotides, or relatively short chains of nucleotides, with neither
consistent 3'-5' links nor specific base sequencing.
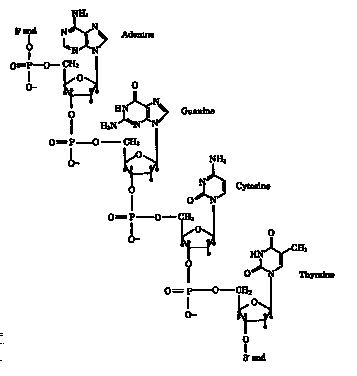
Figure 9-2.
A section from a DNA chain showing the sequence AGCT.
Miller and Orgel summarized their chapter on prebiotic condensation reactions
by saying:
This chapter has probably been confusing to the reader. We believe
that is because of the limited progress that has been made in the study
of prebiotic condensation. Many interesting scraps of information are available,
but no correct pathways have yet been discovered.46
The situation is much the same today.
Summary Discussion of Experimental Results
There is an impressive contrast between the considerable success in synthesizing
amino acids and the consistent failure to synthesize protein and DNA. We
believe the reason is the large difference in the magnitude of the configurational
entropy work required. Amino acids are quite simple compared to protein,
and one might reasonably expect to get some yield of amino acids, even where
the chemical reactions that occur do so in a rather random fashion. The
same approach will obviously be far less successful in reproducing complex
protein and DNA molecules where the configurational entropy work term is
a nontrivial portion of the whole. Coupling the energy flow through the
system to do the chemical and thermal entropy work is much easier than doing
the configurational entropy work. The uniform failure in literally thousands
of experimental attempts to synthesize protein or DNA under even questionable
prebiotic conditions is a monument to the difficulty in achieving a high
degree of information content, or specified complexity from the undirected
flow of energy through a system.
We must not forget that the total work to create a living system goes far
beyond the work to create DNA and protein discussed in this chapter. As
we stated before, a minimum of 20-40 proteins as well as DNA and RNA are
required to make even a simple replicating system. The lack of known energy-coupling
means to do the configurational entropy work required to make DNA and protein
is many times more crucial in making a living system. As a result, appeals
to chance for this most difficult problem still appear in the literature
in spite of the fact that calculations give staggeringly low probabilities,
even on the scale of 5 billion years. Either the work---especially the organizational
work---was coupled to the flow of energy in some way not yet understood,
or else it truly was a miracle.
Summary of Thermodynamics Discussion
Throughout Chapters 7-9 we have analyzed the problems of complexity and
the origin of life from a thermodynamic point of view. Our reason for doing
this is the common notion in the scientific literature today on the origin
of life that an open system with energy and mass flow is a priori
a sufficient explanation for the complexity of life. We have examined the
validity of such an open and constrained system. We found it to be a reasonable
explanation for doing the chemical and thermal entropy work, but clearly
inadequate to account for the configurational entropy work of coding (not
to mention the sorting and selecting work). We have noted the need for some
sort of coupling mechanism. Without it, there is no way to convert the negative
entropy associated with energy flow into negative entropy associated with
configurational entropy and the corresponding information. Is it reasonable
to believe such a "hidden" coupling mechanism will be found in
the future that can play this crucial role of a template, metabolic motor,
etc., directing the flow of energy in such a way as to create new information?
References
1. Albert L. Lehninger, 1970. Biochemistry. New York: Worth
Publishers, p.782.
2. H.P. Yockey, 1977. J. Theoret. Biol. 67, 377; R.W. Kaplan,
1974. Rad. Environ. Biophys. 10, 31.
3. M. Eigen, 1971. Die Naturwiss. 58, 465.
4. G. Steinman, 1967. Arch. Biochem. Biophys. 121, 533.
5. A.G. Cairns-Smith, 1971. The Life Puzzle. Edinburgh: Oliver
and Boyd.
6. F. Crick, 1966. Of Molecules and Men. Seattle: University
of Washington Press, p. 6-7.
7. Eigen, Die Naturwiss., p. 465; S.L. Miller and L.E. Orgel,
1974. The Origins of Life on the Earth. Englewood Cliffs, New
Jersey: Prentice Hall.
8. J.B.S. Haldane, 1965. In The Origins of Prebiological Systems and
of Their Molecular Matrices, ed. S.W. Fox. New York: Academic Press,
p.11.
9. T. Dobzhansky, 1965. In The Origins of Prebiological Systems and
of Their Molecular Matrices, p.310.
10. Ludwig von Bertalanffy, 1967. Robots, Men and Minds. New
York: George Braziller, p.82.
11. G. Steinman and M. Cole, 1967. Proc. Nat. Acad. Sci. U.S. 58,
735; Steinman, Arch. Biochem. Biophys. , p.533.
12. A. Katchalsky, 1973. Die Naturwiss. 60,215; M. Calvin,
1975. Amer. Sci. 63, 169; C.E. Folsome, 1979. The
Origin of Life. San Francisco: W.H. Freeman, p.104; K. Dose, 1983.
Naturwiss. 70, 378.
13. Steinman, Arch. Biochem. Biophys. 121, 533; Steinman
and Cole, Proc. Nat. Acad. Sci. U.S. 5, p.735.
14. H.P. Yockey, 1981. J. Theoret. Biol 91, 13.
15. Katchalsky, Die Naturwiss., p.215.
16. H.R. Hulett, 1969. J. Theoret. Biol 24, 56.
17. Katchalsky, Die Naturwisa., p.216.
18. A.E. Wilder-Smith, 1970. The Creation of Life. Wheaton,
Ill.: Harold Shaw, p.67.
19. G. Nicolis and I. Prigogine, 1977. Self Organization in Nonequilibrium
Systems. New York: Wiley.
20. I. Prigogine, G. Nicolis, and A. Babloyantz, 1972. Physics Today
, p.23-31.
21. Eigen, Die Naturwiss., p.465.
22. Prigogine, Nicolis, and Babloyantz, Physics Today, p.23-31.
23. Ibid; Nicolis and Prigogine, Self Organization in Nonequilibrium
Systems.
24. J.C. Walton, 1977. Origins, 4, 16.
25. P.T. Mora, 1965. In The Origins of Prebiological Systems and of
Their Molecular Matrices, p.39.
26. E.R. Harrison, 1969. In Hierarchical Structures. ed. L.L.
Whyte, A.G. Wilson, and D. Wilson, New York: Elsevier, p.87.
27. Nicolis and Prigogine, Self Organization in Nonequilibrium Systems.
28. Eigen, Die Naturwiss., p.465; 1971. Quart. Rev. Biophys. 4,
149.
29. Kaplan, Rad. Environ. Biophysics, p.31.
30. J. Brooks and G. Shaw, 1973. Origin and Development of Living
Systems. New York: Academic Press, p.209.
31. S.W. Fox and K. Dose, 1977. Molecular Evolution and the Origin
of Life. New York: Marcel Dekker.
32. P.A. Temussi, L. Paolillo, L. Ferrera, L. Benedetti, and S. Andini,
1976. J. Mol Evol 7, 105.
33. S.L. Miller and L.E. Orgel, 1974. The Origins of Life on Earth
Englewood Cliffs, New Jersey: Fn. p. 144.
34. C.E. Folsome, 1979. The Origin of Life. San Francisco:
W.H. Freeman, p.87.
35. Temussi, Paolillo, Ferrera, Benedetti, and Andini, J. Mol. Evol., p.105.
36. K. Bahadur and S. Ranganayaki, 1958. Proc. Nat. Acad. Sci. (India)
27A, 292.
37. S.W. Fox and K. Dose, 1972. Molecular Evolution and the Origin
of Life. San Francisco: W.H. Freeman, p.142.
38. J. Hulshof and C. Ponnamperuma, 1976. Origins of Life 7,
197.
39. C.N. Matthews and R.E. Moser, 1966. Proc. Nat. Acad. Sci. U.S.
56, 1087; C.N. Matthews, 1975. Origins of Life
6, 155; C. Matthews, J. Nelson, P. Varma, and R. Minard, 1977. Science
198 622; C.N. Matthews, 1982. Origins of Life 12,
281.
40. C.N. Matthews, and R.E. Moser, 1967. Nature 215,1230.
41. J.P. Ferris, D.B. Donner, and A.P. Lobo, 1973. J. Mol Biol. 74,
499.
42. J.P. Ferris, 1979. Science 203, 1135.
43. C.N. Matthews, 1979. Science 203, 1136.
44. Katchalsky, Die Naturwiss., p.215.
45. R.E. Dickerson, September 1978. Sci. Amer., p.70.
46. Miller and Orgel, The Origins of Life on the Earth, p.148.
Chapter 8
Chapter 7
Contents
Book Order Form
Scanned and edited for HTML by Lambert Dolphin, June 21, 1996. Not all web
browsers will support the equations and graphics in this paper. For the
most part HTML 2.0 has been used and the results tested on Netscape 2.02.
Footnotes have been interspersed with the text and indented.
Email: Corrections and changes
c in eq. 8-7)].
![]() G2 0 and |
G2 0 and | ![]() G2
|
G2
| ![]() |G1 | so that
|G1 | so that ![]() G1 +
G1 + ![]() G2
=
G2
= ![]() G3
G3 ![]() 0.
0.![]() G = 300 kcal/mole for 100 amino acids),
some investigators have attempted to begin with energy-rich precursors such
as HCN and form polypeptides directly, a scheme which is "downhill"
energetically, i.e.,
G = 300 kcal/mole for 100 amino acids),
some investigators have attempted to begin with energy-rich precursors such
as HCN and form polypeptides directly, a scheme which is "downhill"
energetically, i.e., ![]() G < 0. There are advantages to such an approach; namely,
there is no chemical work to be done since the bonding energy actually decreases
as the energy-rich precursors react to form more complex molecules. This
decrease in bonding energy will drive the reaction forward, effectively
doing the thermal entropy work as well. The fly in the ointment, however,
is that the configurational entropy work is enormous in going from simple
molecules (e.g., HCN) directly to complex polymers in a single step (without
forming intermediate biomonomers).
G < 0. There are advantages to such an approach; namely,
there is no chemical work to be done since the bonding energy actually decreases
as the energy-rich precursors react to form more complex molecules. This
decrease in bonding energy will drive the reaction forward, effectively
doing the thermal entropy work as well. The fly in the ointment, however,
is that the configurational entropy work is enormous in going from simple
molecules (e.g., HCN) directly to complex polymers in a single step (without
forming intermediate biomonomers).
